Warning: Paket 'plyr' wurde unter R Version 4.2.3 erstelltBeautiful plots while simulating loss in two-part Procrustes problem
Today, I was working on a two part Procrustes problem and wanted to find out why my minimization algorithm sometimes does not converge properly or renders unexpected results. The loss function to be minimized is
with
By plugging
When trying to find out why the algorithm to minimize
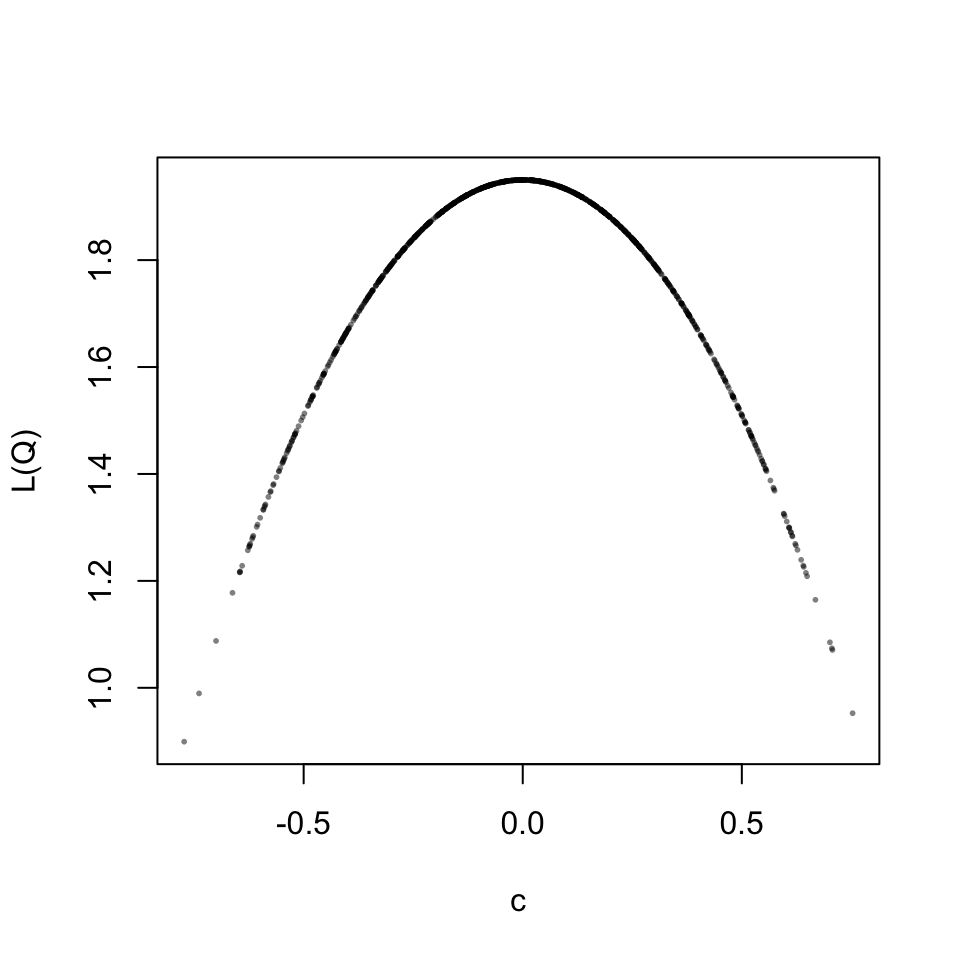
This is a well behaved relation, for each scaling parameter the loss value is identical. Now let’s look at the full two-part loss function. The result turns out to be beautiful.
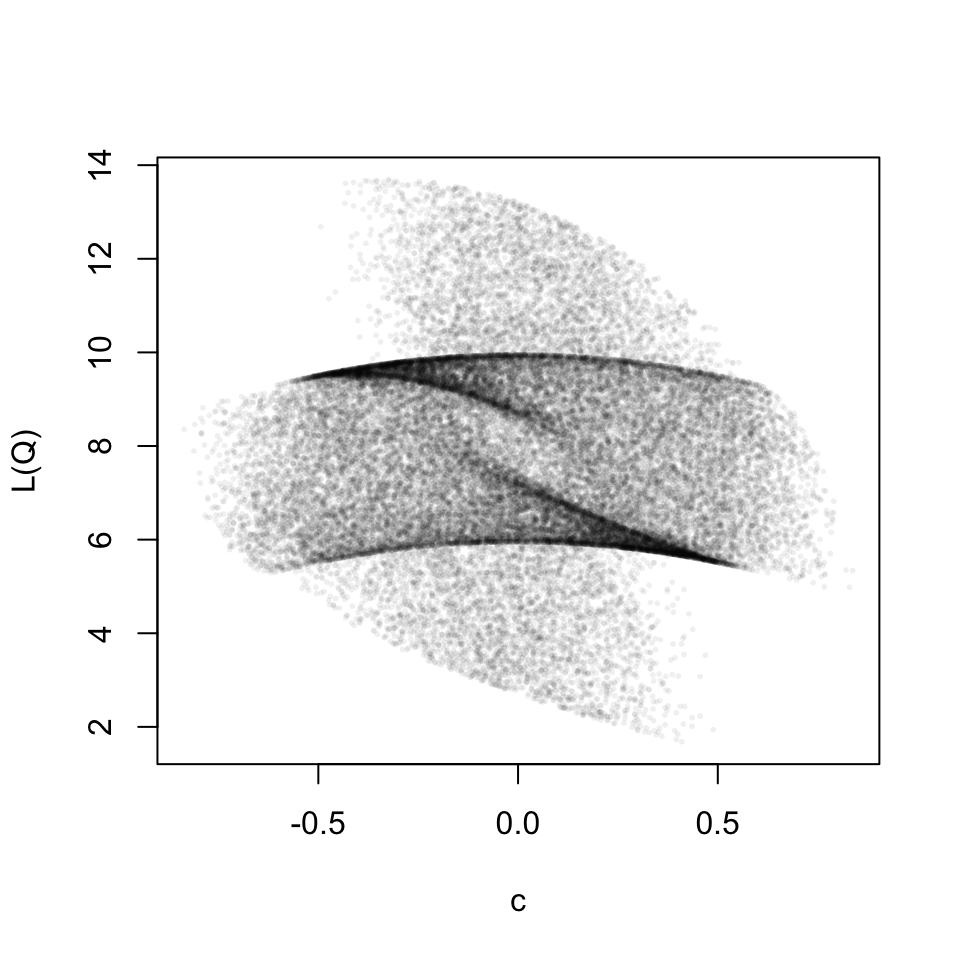
For the graphic above I used the following matrices.
And the following R-code.
tr <- function(X) sum(diag(X))
# random matrix type 1
rmat_1 <- function(n=3, p=3, min=-1, max=1){
matrix(runif(n*p, min, max), ncol=p)
}
# random matrix type 2, sparse
rmat_2 <- function(p=3) {
diag(p)[, sample(1:p, p)]
}
# generate random rotation matrix Q. Based on Q find
# optimal scaling factor c and calculate loss function value
#
one_sample <- function(n=2, p=2)
{
Q <- mixAK::rRotationMatrix(n=1, dim=p) %*% # random rotation matrix
diag(sample(c(-1,1), p, rep=T)) # additionally
s <- tr( t(Q) %*% t(A1) %*% B1 ) / norm(A1, "F")^2 # scaling factor c
rss <- norm(s*A1 %*% Q - B1, "F")^2 +
norm(A2 %*% Q - B2, "F")^2
c(s=s, rss=rss)
}
# find c and rss or many random rotation matrices
#
set.seed(10) # nice case for 3 x 3
n <- 3
p <- 3
A1 <- round(rmat_1(n, p), 1)
B1 <- round(rmat_1(n, p), 1)
A2 <- rmat_2(p)
B2 <- rmat_2(p)
x <- rdply(40000, one_sample(3,3))
plot(x$s, x$rss, pch=16, cex=.4, xlab="c", ylab="L(Q)", col="#00000010")Below you find some more graphics with different matrices as inputs.

Here, we do not have a one-to-one relation between the scaling parameter and the loss function any more. I do not quite know what to make of this yet. But for now I am happy that it has aesthetic value.